Research
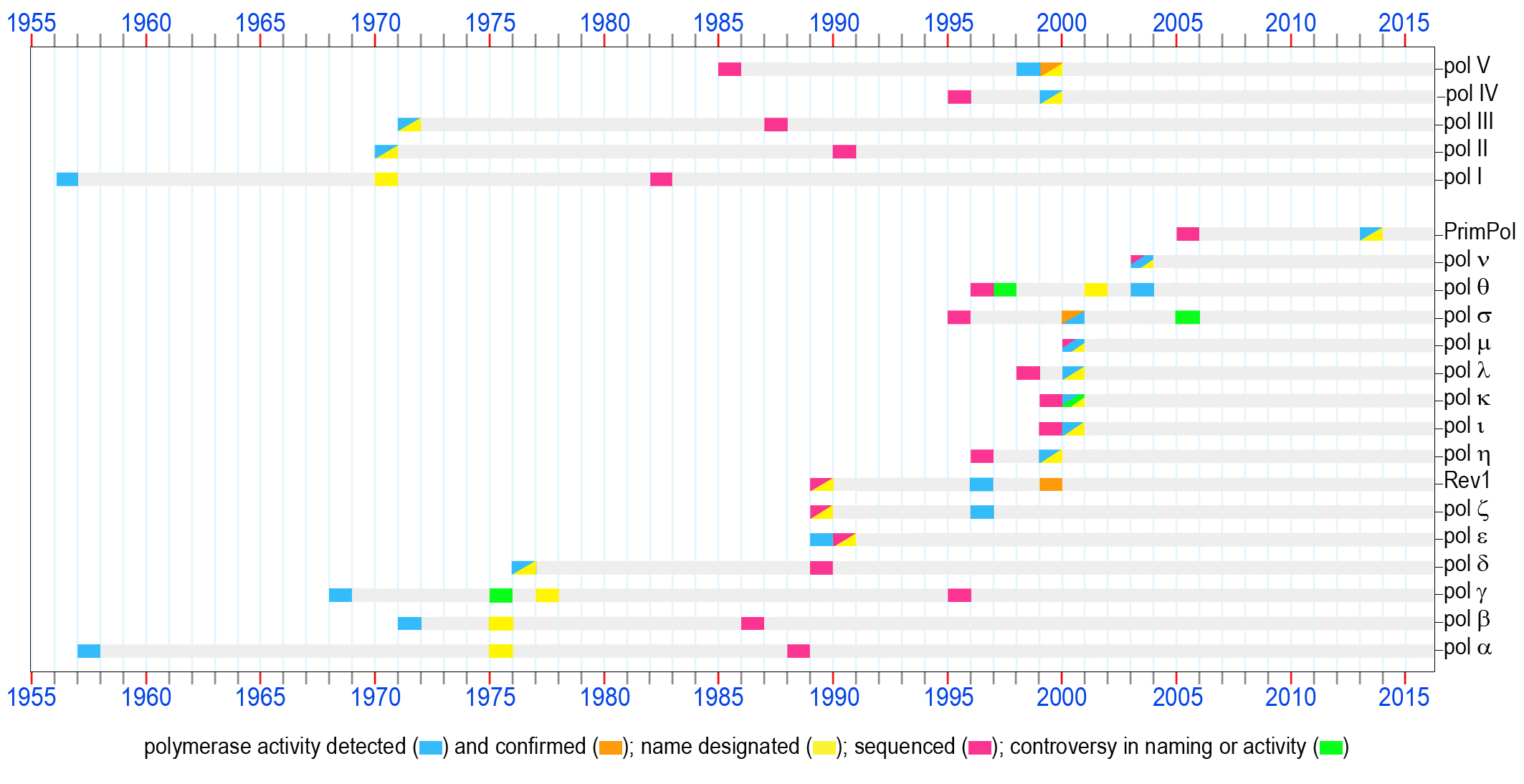
DNA polymerases, 60 years of discoveries;
click on the picture to see a larger version and the descriptionUnder optimal conditions, the fidelity of DNA replication is extremely high. Indeed, it is estimated that, on average, only one error occurs for every 10 billion bases replicated. However, given that living organisms are continually subjected to a variety of endogenous and exogenous DNA–damaging agents, optimal conditions rarely prevail in vivo. Even though all organisms have evolved elaborate repair pathways to deal with such damage, the pathways rarely operate with 100 percent efficiency. As a consequence, cells have developed a mechanism for synthesizing past persisting DNA lesions, a process commonly referred to as translesion DNA synthesis (TLS) or translesion replication (TR). Similar to DNA replication and DNA repair, TLS depends on DNA polymerases each playing a specific role in copying modified genomic sequences.
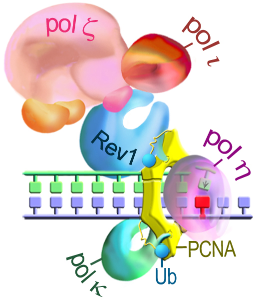
TLS in eukaryotic cells;
click on the picture to see the pathway
Virtually all DNA polymerases characterized in vitro appear able to traverse past damaged DNA sites although with vastly different efficiencies, and fidelities, and often with the help of protein partners. High fidelity replicative polymerases are the least adept to perform this function. In contrast, all organisms contain specialized DNA polymerases which can replicate damaged DNA quite efficiently. Within the TLS polymerase superfamily, there is a group of enzymes that appear to be specifically designed to accomplish the lesion bypass task most effectively. The majority of these enzymes comprise the Y-family of DNA polymerases that are phylogenetically conserved from bacteria to humans. Based on phylogenetic relationships, Y-family polymerases may be broadly classified into five subfamilies: DinB-like (polIV/pol kappa–like) proteins are ubiquitous and found in all domains of life; in contrast, the Rev1-like, Rad30A (pol eta)–like, and Rad30B (pol iota)–like polymerases are found only in eukaryotes; and the UmuC (polV)–like polymerases only in prokaryotes.
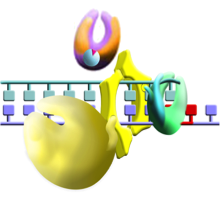
TLS in prokaryotes;
click on the picture to see the pathway
It is clear that many mutations are generated during the cell's attempts to facilitate replication past normally replication-blocking lesions. Our projects aim to understand the molecular events that influence the fidelity of the genome, facilitating both evolution and species stability. We investigate the molecular mechanisms by which mutations are introduced into damaged DNA through TLS in all three kingdoms of life: bacteria, archaea, and eukaryotic cells. The dynamics of mutagenesis are also of interest, given the importance of mutations in genetic diseases, oncogenesis, and developmental abnormalities. Our studies involve a variety of molecular and biochemical techniques, including PCR, protein purification and characterization and yeast/bacterial molecular genetics.
 BACK TO TOP
BACK TO TOP